Specifying the penetrance
A critical aspect affecting the analysis output is the model connecting genetic variant and disease. This is encoded by the penetrance function, which shinyseg allows to build with different degrees of complexity using the Penetrance panel.
Inheritance pattern
The first decision regarding the penetrance relates to the inheritance model. Choose one of the options on the header:
AD: autosomal dominant.AR: autosomal recessive.AI: autosomal incomplete dominance.XD: X-linked dominant.XR: X-linked recessive.XI: X-linked incomplete dominance.
Note that incomplete dominance, where you can specify different
parameters for the carriers of 1 and 2 copies of the variant, will be
unavailable unless you also switch to the Liability class mode. An
example of an analysis using X-linked incomplete dominance can be found
at #2 X-linked inheritance.
Penetrance mode
Next, it is time to assign the probabilities of observing the analysis phenotypes conditional on carriership status, hereafter referred to as penetrances. shinyseg offers two different ways to do it:
- Relative risk, where the penetrances are computed by the app based on certain parameters.
- Liability class, where the user manually specifies them.
The mode can be switched anytime with the input on the left of the
header, but note that Relative risk will be unavailable if incomplete
dominance is selected.
1) Relative risk
This is a parametric version of the survival penetrances described in Belman et al. (2020). It is based on:
- Baseline lifetime risk, mean and SD: the lifetime risk, mean, and standard deviation of disease onset in non-carriers and heterozygous carriers in recessive inheritance.
- Hazard ratios: the relative risks in homo-, hemi-, and heterozygous carriers in dominant inheritance, compared to the baseline. They can be either constant or age-dependent and may also be specified through a variant-associated lifetime risk.
These parameters are entered using a table generated from the phenotypes
specified in the pedigree table. For example, the following describes a
constant relative risk model in which non-carriers (neg) have a
lifetime risk of 1% (phenocopy rate) with onset at 70±15 years of age,
while heterozygous and homozygous carriers (het/hom) have a lifetime
risk of 75%.
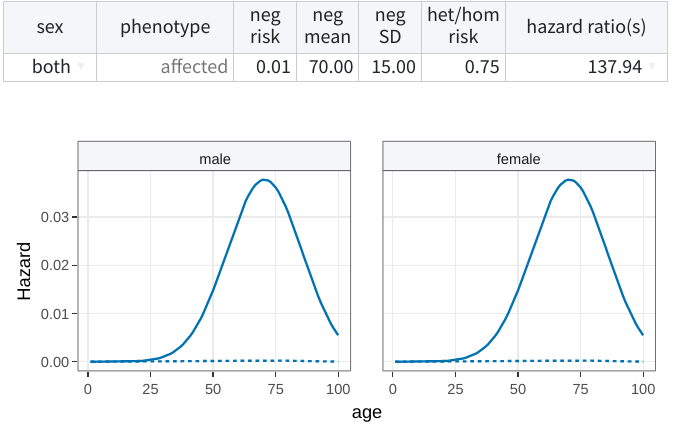
Below the table, you will also find a plot which can serve as a visual aid displaying the cumulative incidences, hazards, or hazard ratios determined by the current selection.
None of the model parameters can be missing.
Sex-specific penetrances
By default, shinyseg generates a unique entry for each phenotype. This
means that the specified penetrances will apply to bothsexes. However,
you can click on this field and select male or female to create a
sex-dependent specification.
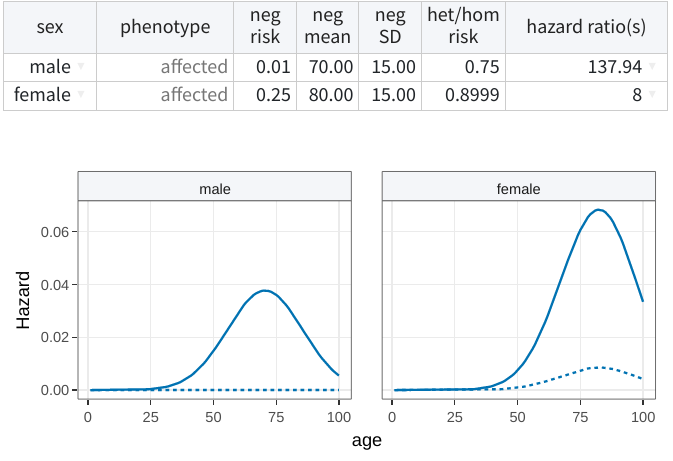
To undo it, change back to both any of two the sex-specific entries.
Hazard ratios
The previous examples ignored the hazard ratio(s) column, which contained a constant (age-independent) relative risk of the disease in variant carriers compared to the baseline. This is because shinyseg allows inputting the variant-associated lifetime risk instead, and updates the other column accordingly. However, users can also manipulate the hazard ratios directly, in which case it will be the other input that is updated.
Although more complicated, the direct specification of the hazard
ratio(s) has the advantage of allowing for age-dependent relative
risks if, instead of a single value, a vector is provided. Specifically,
it admits up to 10 comma-separated values representing, loosely
speaking, the hazard ratios at evenly spaced ages from 1 to 100 years.
For instance, 1,10,10,20,10 would roughly correspond to the values at
ages 1, 25, 50, 75 and 100. The app will interpolate/smooth these across
the lifespan using a B-spline basis — you may adjust the smoothing
degree with the Splines input at the bottom. The examples #3 Two
phenotypes and #4 Breast cancer and
BRCA1 showcase penetrance models with age-dependent
relative risks.
If you misspecify the hazard ratio(s), their values will revert to the previous ones to prevent issues.
Note that it is also possible to leverage the variant-associated
lifetime risk, as earlier, but combined with a vectorial specification
of the hazard ratios. For instance, changing the hazard ratio(s) to
1,2,2,1 defines a model where the relative risk is higher at middle
ages. Subsequently, resetting the lifetime risk to 75% will scale these
values accordingly, resulting in:
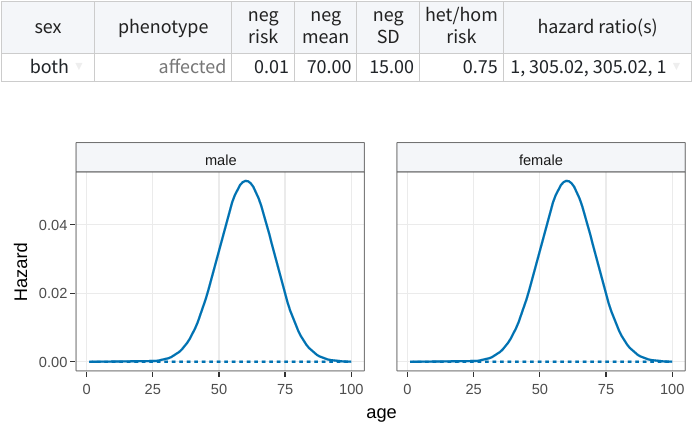
Optimal parameters
As choosing the model parameters can be challenging, shinyseg also offers an Assistant to optimize them based on user-provided cumulative incidence data.
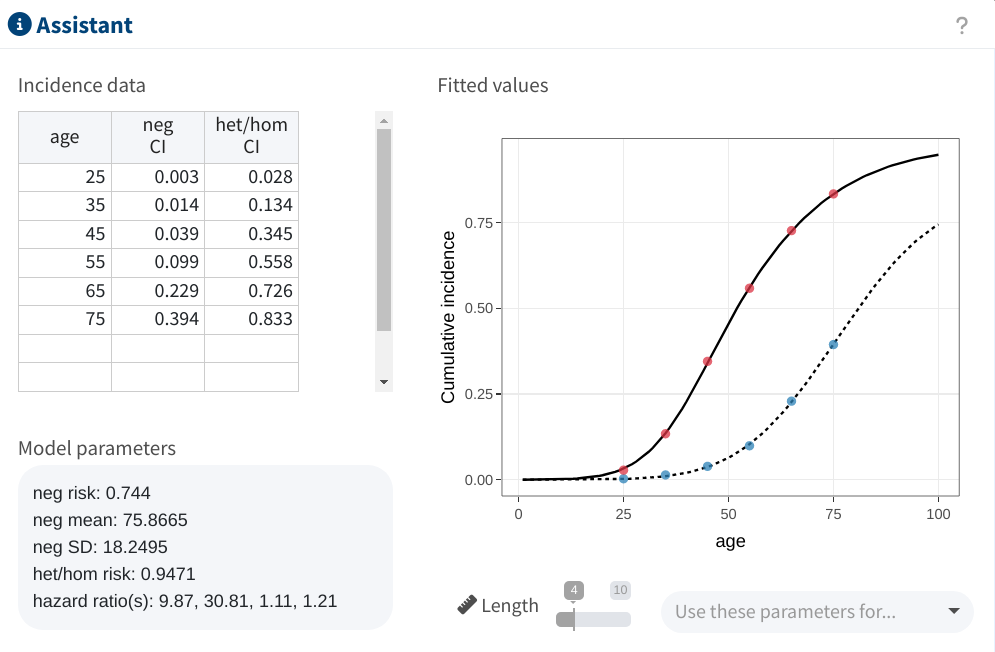
To use it, simply specify pairs of age (integer between 1-100) and their corresponding cumulative incidence in the table provided. The Length slider also allows to fine tune the model’s complexity by defining the number of values for the hazard ratio vector. The app will then find the optimal parameters, which you may easily transfer to your analysis with Use these parameters for….
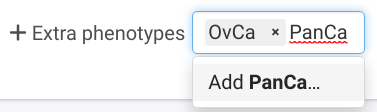
Extra phenotypes
Sometimes, as in #4 Breast cancer and BRCA1, not all phenotypes relevant for the penetrance function may be observed in the analysis families. This means that, by default, they will not appear in the table where you can set their parameters. To include them, type their names in the Extra phenotypes input located at the bottom of the Penetrance panel.
2) Liability class
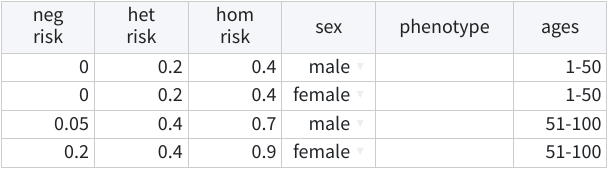
Alternatively, users can manually define penetrances using a table. This table contains 2 (dominant/recessive) to 3 (incomplete inheritance) columns with the penetrances themselves, followed by an additional 3 columns specifying the classes they apply to:
- sex:
male,female, orbothfor classes referring to males, females, or both sexes, respectively; leaving this cell empty is equivalent to specifyingboth. - phenotype: a free-text field whose values should match the
phenotypes in the pedigree table. Note that here,
nonaffis treated as another phenotype whose classes must be specified (unless there are no unaffected cases); leaving this cell empty indicates that the class applies to all phenotypes. - ages: an integer from 1 to 100 or ranges between these values (e.g., 10-20, 30-100…) indicating the ages to which the class applies; leaving this cell empty is equivalent to specifying ages 1-100.
For instance, the following illustrates an autosomal dominant model with no phenocopies, where both heterozygous and homozygous carriers have a 100% chance of disease onset:
And the following defines a sex- and age-dependent specification with incomplete dominance:
The table may be created interactively or by uploading a file via Load file. If opting for the latter, shinyseg expects a tabular file without column headers and a structure resembling the one shown in-app.
There should be no sex/phenotype/age overlaps. All family members should map to a single liability class without missing penetrances.